Research interests
Here is a list of the research projects I am currently working on:
Also I have been involved with over the years in other projects, which I still like to discuss or be updated:
1) DNA damage: reactivity and dynamical aspects by hybrid QM/MM-MD simulations.
a) Oxidative DNA damage, singlet-oxygen reactivity
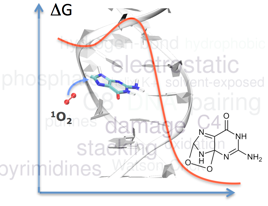
We explore by multiscale simulations key reactions unperpinning oxidatively-induced DNA damage.
QM/MM-MD free energy profiles are obtained with fDynamo, CPMD... which corroborate mechanistic pathways (accessible free energy barriers and
exergonic processes). They can differ from the one at the nucleobase level, reversing nucleobase selectivity [1].
The methodologies we employ are close to the ones used in enzymatic catalysis.
We can compare the reactivity of nucleobases [2] and get insights into the structural footprint of a lesion, given that
no major structural distortion comes into play.

Further references:
- "What singles out the G[8-5]C intrastrand DNA cross-link ? Mechanistic and structural insights from QM/MM simulations.", C. Patel, J. Garrec, C. Dupont and E. Dumont, Biochemistry, 2013, 52, 425-431 doi.
- "Insights into the formation of oxidative intrastrand cross-link lesions of DNA from QM/MM molecular dynamics simulations.", J. Garrec,
C. Patel, U. Rothlisberger and E. Dumont, J. Am. Chem. Soc, 2012, 134, 2111-2119. doi.
- "Probing the reactivity of singlet oxygen with purines.", E. Dumont, R. Gruber, E. Bignon, C. Morell, Y. Moreau, A. Monari, J.-L. Ravanat, Nucl. Acids Res., 2016, 44:56-62
- "Singlet oxygen guanine onto guanine: reactivity and structural signature within the B-DNA helix", E. Dumont, R. Grüber, E. Bignon, C. Morell, J. Aranda, J.-L. Ravanat, I. Tuñón, Chem. Eur. J., 2016
b) Structural outcome of defect-containing oligonucleotides upon MD investigation
We also investigate dynamics up to the microsecond range for complex DNA lesions such as intra- or interstrand cross-links, multiply damaged sites.
We derive force field parameters for DNA defects, and perform classical MD simulations to generate in silico the 3D structures upon induction of
complex DNA damage. This information concerns the bending of the helix, the more or less pronounced disruption of Watson-Crick pairing and shed light on possible structural reasons for a lack of repair.

Further references:
- "What singles out the G[8-5]C intrastrand DNA cross-link ? Mechanistic and structural insights from QM/MM simulations.", C. Patel, J. Garrec, C. Dupont and E. Dumont, Biochemistry, 2013, 52, 425-431 doi.
2) Excited states and photosensitization


We delineate energies coming into play in photosensitization process, such as triplet transfer energy or hydrogen abstraction.
This necessitates to first assess binding modes, which are often competitive. Then the energy transfer process or generation
of intermediates species such as singlet oxygen can be described relying on DFT or CASPT2 methods.
Further references:
- "Benzophenone and DNA: Evidences for a Double Insertion Mode and its Spectral Signature." E. Dumont and A. Monari, J. Phys. Chem.
Lett., 2013, 4:4119-4124 link
- "Hydrogen abstraction by photoexcited benzophenone: consequences for DNA photosensitization." M. Marazzi, M. Wibowo, H. Gattuso, E. Dumont, D. Roca-Sanjuan, A. Monari, Phys. Chem. Chem. Phys., 2016, 18, 7829-7836
- "Resolving the Benzophenone DNA-Photosensitization Mechanism at QM/MM Level."E. Dumont, M. Wibo, D. Roca-Sanjuan, M. Garavelli, X. Assfeld, A. Monari, J. Phys. Chem. Lett, 2015, 6:576-580
- "DNA photosensitization by an "insider". Photophysics and triplet energy-transfer of 5-methyl-2-pyrimidone deoxyribonucleoside.", E. Bignon, H. Gattuso, C. Morell, E. Dumont, A. Monari, Chem. Eur. J., 2015
3) Interaction peptides-inorganic systems
We also investigate dynamics at microsecond range for proteins--lanthanides or hybrid polyoxometalates,
in close collaboration with experimentalists (coll. O. Maury and E. Lacote). We characterize and assess the free energies of interactions using MM/PBSA or DFT calculations: comparison with NMR is crucial to validate our computational approach.

Further references:
- "Exploration of the supramolecular interactions involving tris-dipicolinate lanthanide complexes in protein crystals by a combined biostructural, computational and NMR study" E. Dumont, G. Pompidor, A. D'Aléo, J. Vicat, L. Toupet, R. Kahn, E. Girard, O. Maury and N. Giraud, Phys. Chem. Chem. Phys, 2013, 15, 18235-18242 link
- "Grafting of Secondary Diol-Amides onto [P2W15V3O62]$9-] Generates Hybrid Heteropoly Acids."
D. Lachkar, D. Vilona, E. Dumont, M. Lelli, E. Lacote, Ang. Chem. Int. Ed.}, 2016, accepted
4) Radical chemistry
Many systems we are interested in deals with radical, notably for oxidatively-induced DNA lesions
but also in the case of π-dimers tailored for supramolecular interactions (coll. ENSL, C. B\"ucher).

5) DFT benchmarking
The biomolecules we are investigating calls for modern functionals, which we often have to benchmark against
post Hartree-Fock calculations or eventually calibrate against experimental spectra: indeed, our calculations
implied triplet states, radical species, charge transfer...
Sometimes, it constitutes a work on its own.
Further references:
- "Stability of the Guanine Endoperoxide Intermediate: A Computational Challenge for Density Functional Theory",
R. Gr\"uber, A. Monari, E. Dumont, J. Phys. Chem. A, 2014, 118:11612-11619
doi.
- "Superior Performance of Range-Separated Hybrid Functionals for Describing disulfide radical anions and similar systems", C. Dupont, E. Dumont, and D. Jacquemin, J. Phys. Chem. A, 2012, 116:3237-3246 doi.
- "Towards an accurate treatment of σ* σ transitions: moving on to N6.- E. Dumont, N. Ferre and A. Monari, Chem. Phys. Lett., 2013, 580:14-20 link
- "Improved DFT description of intrastrand cross-link formation by inclusion of London dispersion corrections." C. Dupont, C. Patel and E. Dumont, J. Phys. Chem. B, 115, 15138-15144 doi.
- "Performances of recently-proposed functionals for describing disulfide radical anions and similar systems", E. Dumont, A. D.
Laurent, X. Assfeld and D. Jacquemin, Chem. Phys. Lett., 2011, 501, 245-251.
doi
Last modification : May 2016
|





