[ Oritatami systems | Download | Examples of .os files | File format ] Oritatami systems
The Oritatami model was introduced in 2016 in (GMSS, in IJMS 2019) to study the computational potential of RNA cotranscriptional folding as first shown in wet-lab experiments by (GRA, in Science 2014).
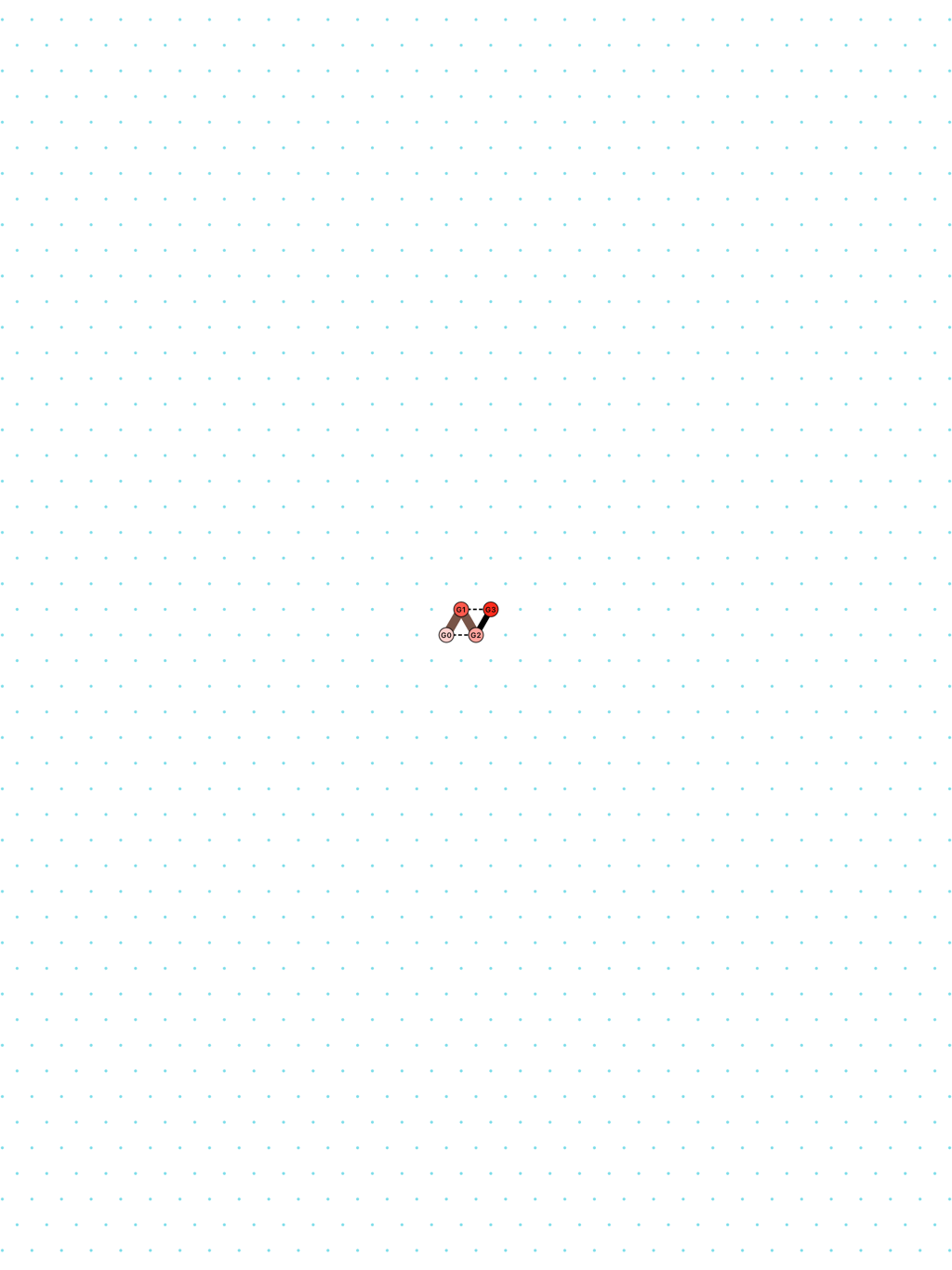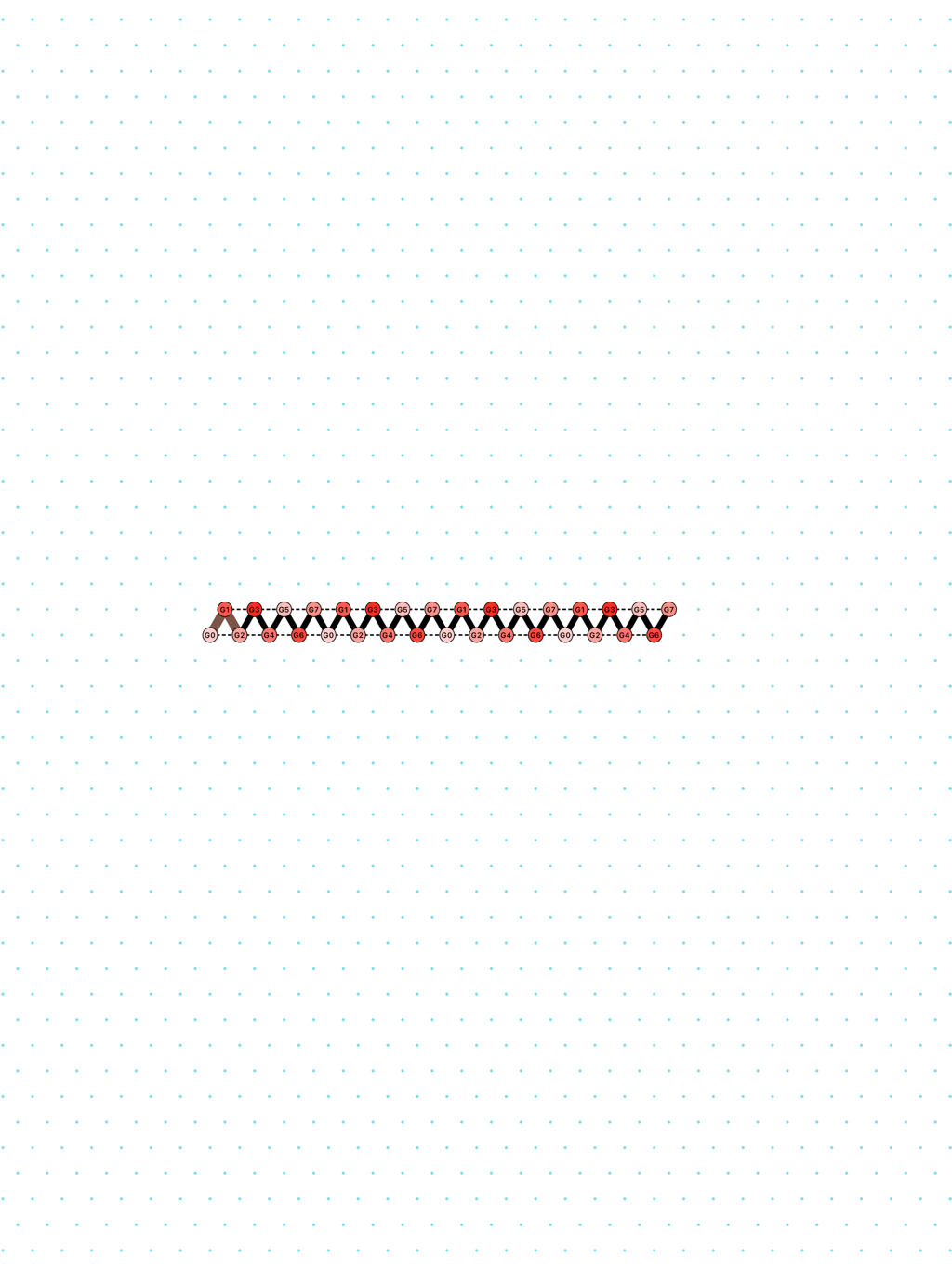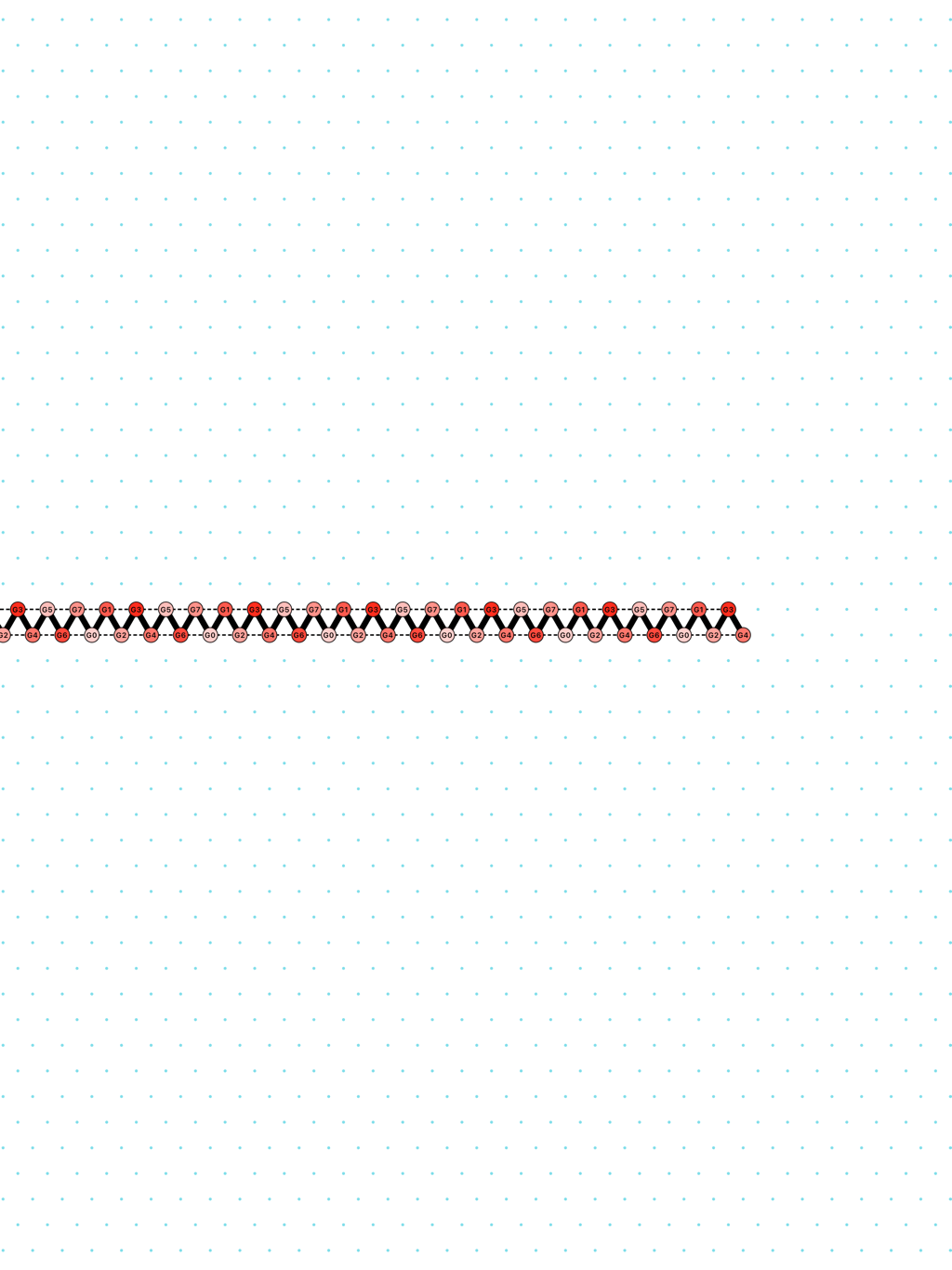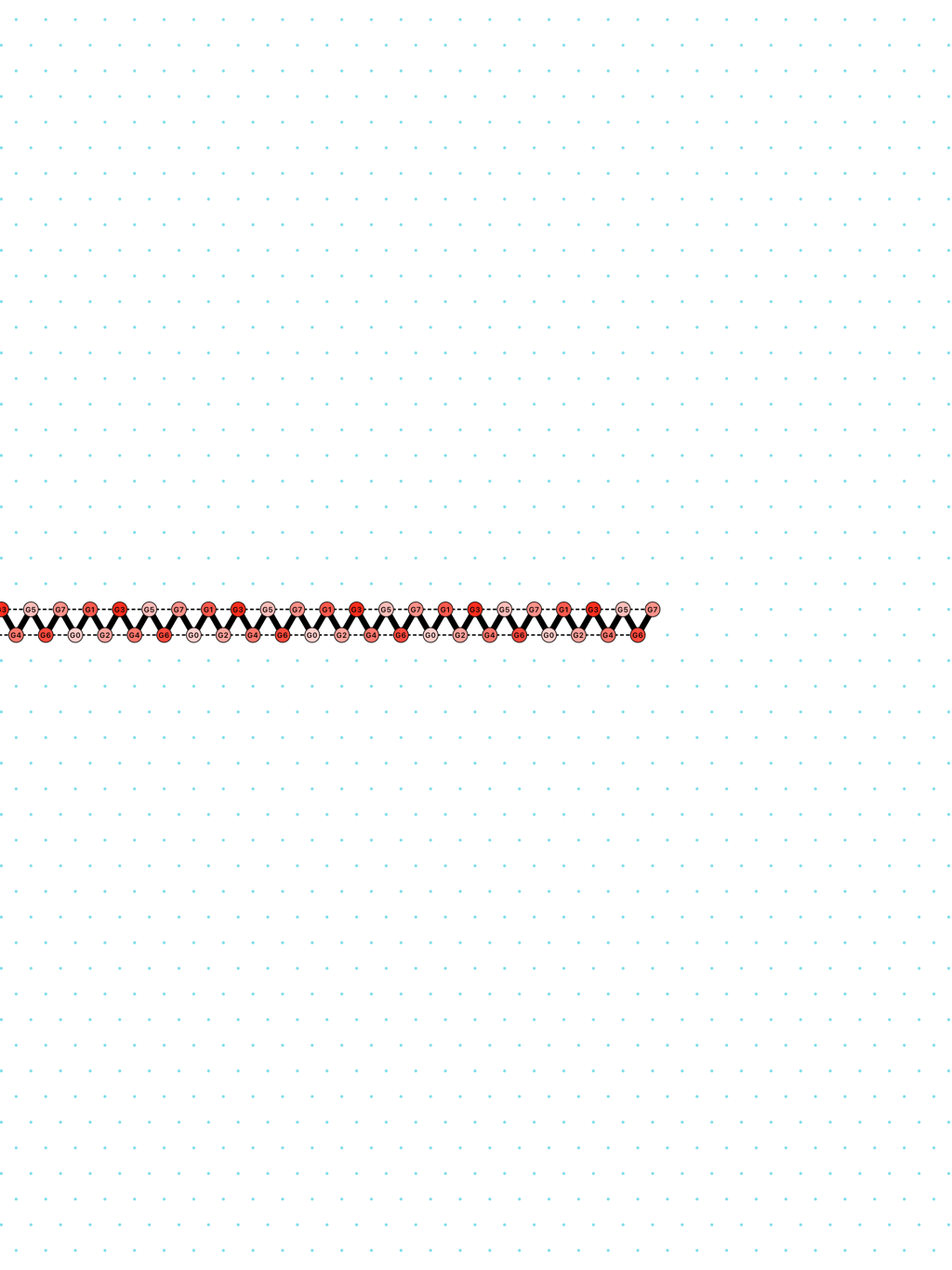
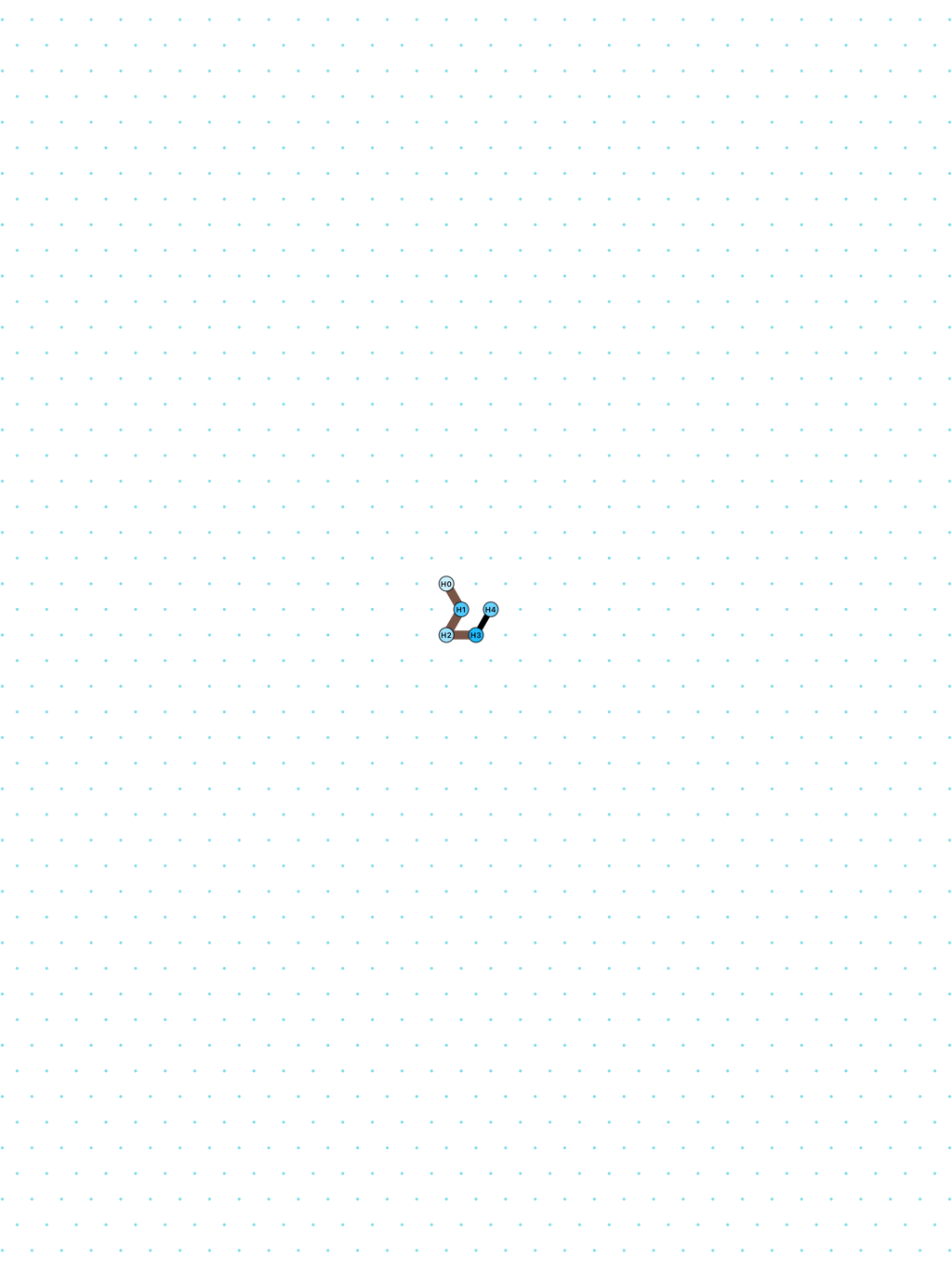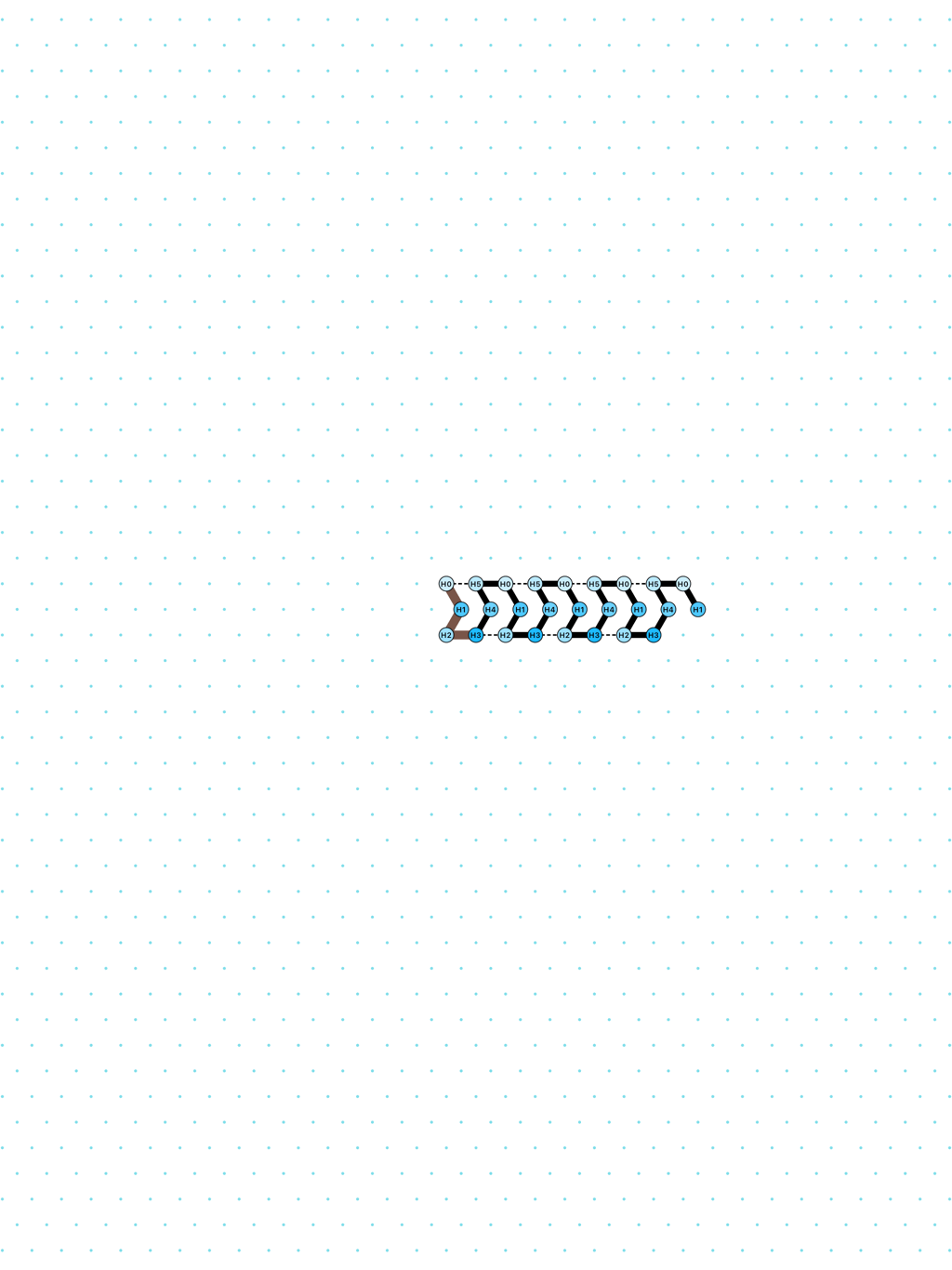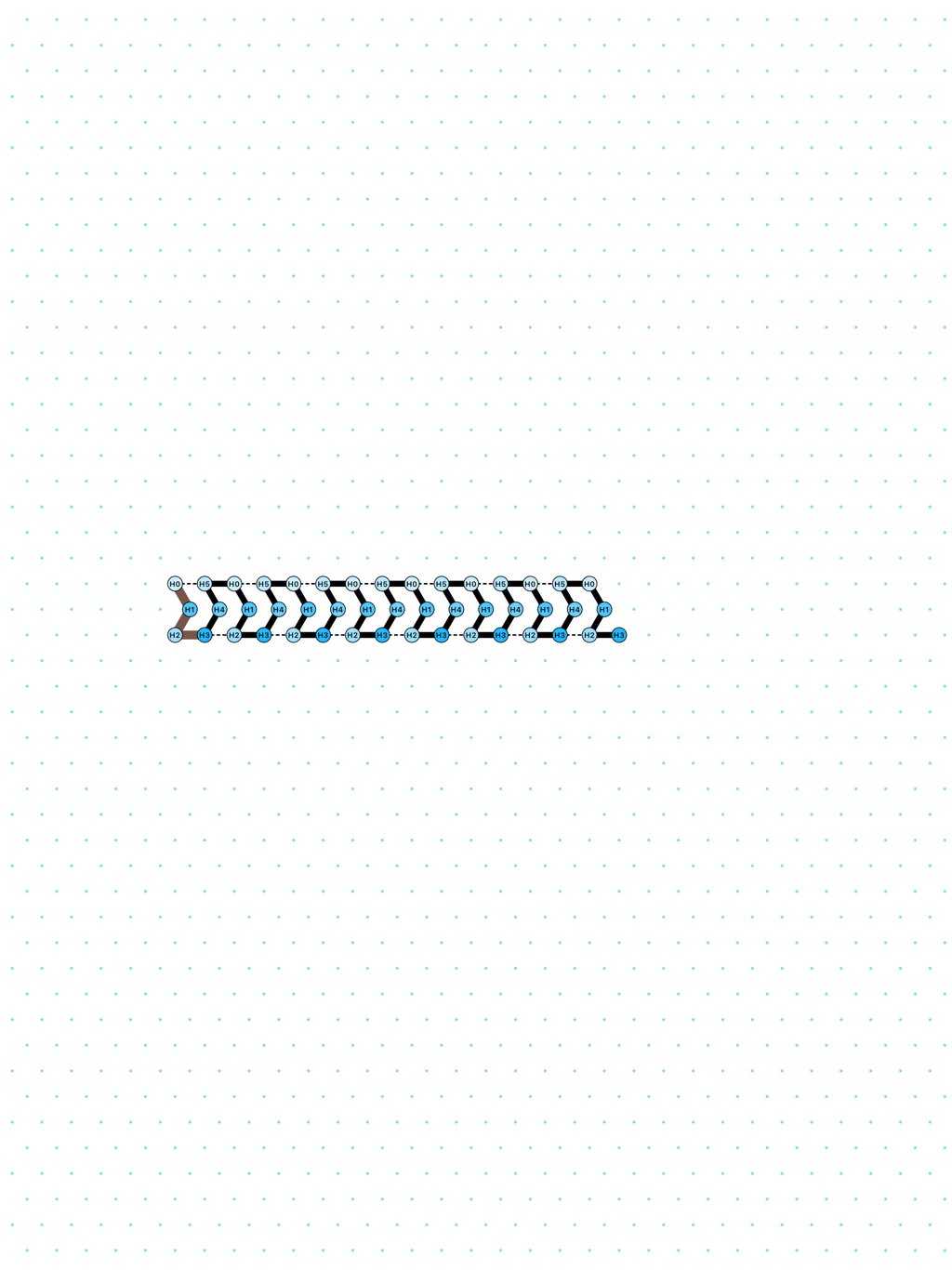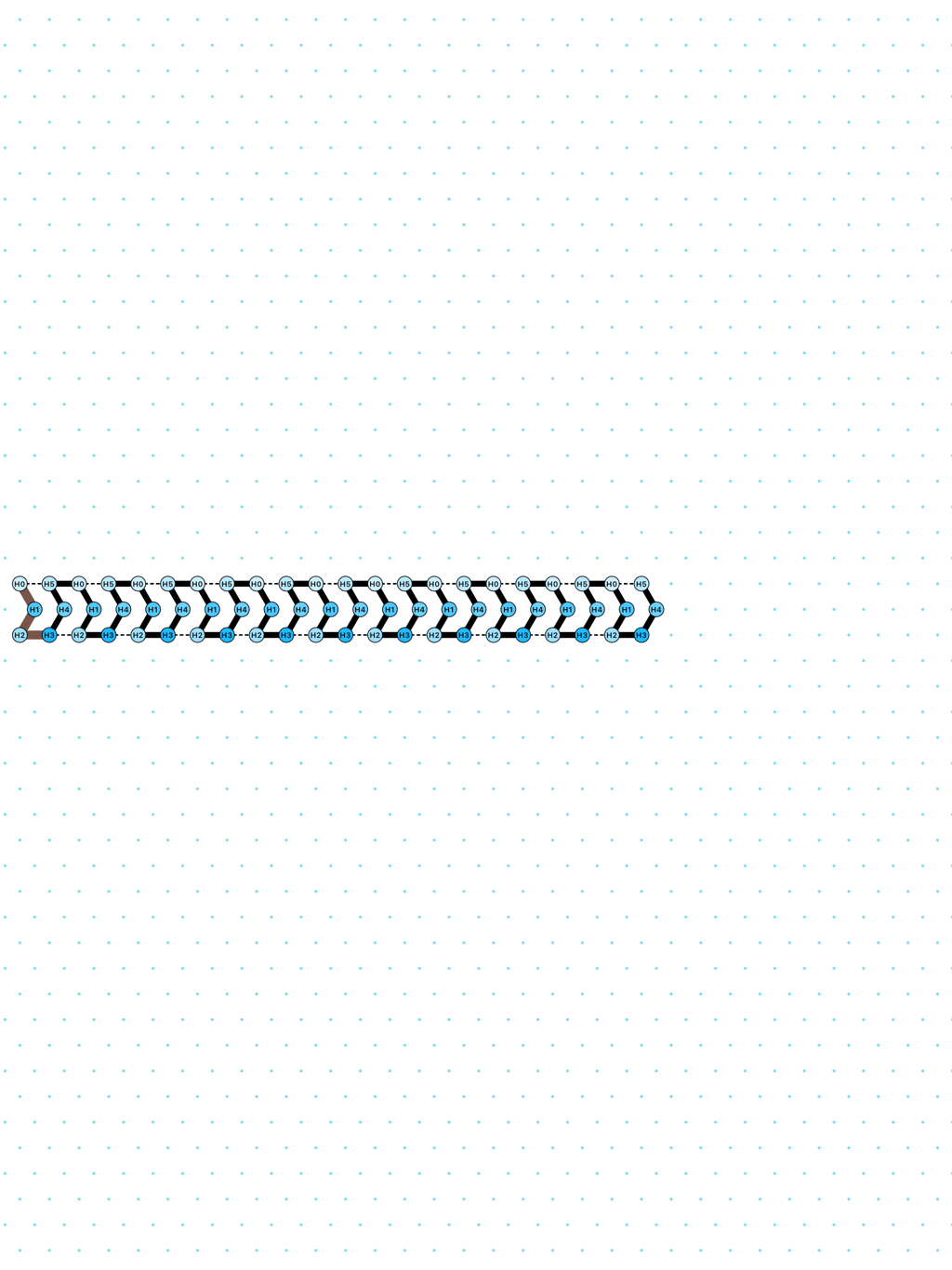
In Oritatami systems (OS), we consider a finite set of bead types, and a periodic sequence of beads, each of a specific bead type. Beads are attracted to each other according to a fixed symmetric relation. In any folding, a bond is formed between any pair of beads located at adjacent positions and attracting each other. At each step, the δ latest nascent beads in the sequence are allowed to explore all possible positions, and adopt only those positions that minimise the energy, or otherwise put, those positions that maximise the number of bonds in the folding. δ is called the delay. ``Beads'' are a metaphor for domains, i.e. subsequences, in RNA and DNA (and are thus not limited to 4 types only). The Oritatami model has proven to be a fruitful framework to study molecular reconfigurability, one of the most promising directions to reduce error in wetlab molecular implementation as error might be erased by reconfiguration later on.
Previous work on Oritatami includes among others the implementation of a binary counter (GMSS, in IJMS 2019), the Heighway dragon fractal (MSU, in Automata 2018), folding of shapes at small scale (DHOMRSST, in DNA 2018), NP-hardness of the rule minimization (HK, in TCS 2019; OS, in TCS 2017), a study of its parameters (RS, in FI 2017), a polynomial-time Turing machine simulation (GMSS, in ISAAC 2018), and a 1D cellular automata intrinsic simulation (PSSU, in LATIN 2020).
Download OS simulator
OS simulator is an iPad app that runs arbitrary oritatami systems efficiently. Oritatami systems have to be loaded into the app as a .os file, whose json format is described below.
OS simulator can be downloaded directly from the iOS Appstore. Its source code is also available:
source code: OSsimulator.zip
Examples of .os files
The following .os files were built using the OS python3 class in OS_class.py. These .os files should be placed in the OS Simulator folder of the iOS Files app or sent to the ipad by Airdrop to be opened by OS simulator.
Download all the examples: examples.zip
a delay-2 glider: [
glider_2.py|Glider_2.os]
a delay-3 glider: [
glider_3.py|Glider_3.os]
an alternation of two types of delay-3 gliders: [
glider_2+3.py|Glider_2+3.os]
a delay-3 switchback: [
switchback_3.py|Switchback_3_k3_p6.os]
The .os file format
The beadtype names are expected to adopt the format "CCxx" where "CC" is a non-numeric string of arbitrary length and "xx" is an arbitrary integer. The non-numeric part "CC" is referred as the category of the bead type and the number "xx" as its number in this category. Beadtype with the same category are colored by defaults with a different shade of the category color. Note that beadtypes do not need to be declared and are created as they are discovered when reading the .os file.
We recommand to use the OS python3 class in OS_class.py to build an .os file. (see examples above)
The .os file adopts the json format with the following fields:
"name": a string that contains the display name of the OS"delay": a positive integer that is the OS delay"categoryColors": a dictionnary (see below)"rule": an array of array of pair of strings for the names of attracting beadtypes"seedConformation": an array of strings encoding an alternation of beadtype names and directions ("NW", "NE", "E", "SE", "SW" or "W") starting and ending with a beadtype"compactTranscriptPrefix": ajson-array encoding of the prefix of the transcript"compactTranscriptPeriod": thejson-array encoding of the period of the transcript"periodCount": an non-negative integer for the number of times the period of the transcript is repeated (0for infinite)
The "categoryColors" field expects a json-encoded dictionnary associating each category name with a color json-encoded as either:
"rgba": an array of up to 4 floats for the red, green, blue and alpha components (the default value for missing components is1if the array is too short)"hsba": an array of up to 4 floats for the hue, saturation, brightness and alpha components (the default value for missing components is1if the array is too short)"name": a string for the name of a material color (e.g."red500")
The "compactTranscriptPrefix" and "compactTranscriptPeriod" fields expect a json-array encoding the bead type sequences as a datastructure <S> defined recursively as follows:
"CCxx": a string encoding a beadtype name"XXXX": a string "XXXX" standing for the beadtype sequence declared as "XXXX" earlier or the bead type name "XXXX0" (with index 0) if "XXXX" was not declared earlier[<S1>,...,<Sk>]: an array encoding the concatenation of the beadtype sequences encoded by each<Si>["unfold", i, j, <S1>,...,<Sk>]: an array encoding the subsequence of bead types indexed fromitoj-1in the infinite cyclic concatenation of the expansions of the encodings<S1>,...,<Sk>["repeat", i, j, <S0>,...,<Sk-1>]: an array encoding for the concatenation of the expansions of the sequences<Si mod k>,...,<S(j-1) mod k>["declare", "XXXX", <S1>,...,<Sk>]: an array encoding the declaration of a constant name"XXXX"whose value is the concatenation of the sequences encoded by<S1>,...,<Sk>