Research interests
My research interests cover a wide range of topics both in physics (statistical physics, confined polymers, out-of-equilibrium physics) and biophysics (epigenetics, molecular motors, cancer physics, nuclear pore complex, physical virology). Most of these studies are based on single molecule experiments and modeling. The specificity here is to focus on a physical mechanism at stake in a biological phenomenon (e.g. cargo confinement in nuclear pore complex) and to investigate this process by means of controlled in vitro or ex vivo experiments. My current research activity is mostly focused on the transport of biomolecules in artificial and biological nanopores.
Translocation of biomolecules through nanopores
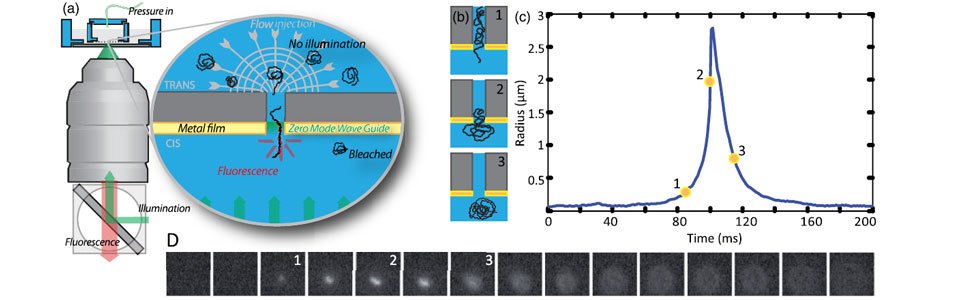
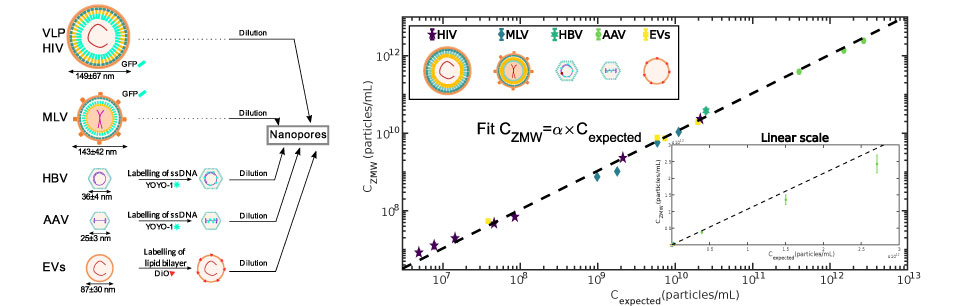
We established a method to follow individual biomolecules such as nucleic acids (DNA, RNA) or proteins (individual or complexes) as they translocate through artificial nanopores. In our case the nanoporous membranes are unexpensive track etch membranes that were originally designed for water filtration. These polycarbonates membranes are evaporated with a thin layer of gold in order to produce Zero-Mode waveguides which are metallic nano-structures at the exit of pore. These structures strongly enhance and localize the fluorescence emission of the biomolecules when they reach the exit of the nanopore. The presence of high densities of nanopores in these membranes natively induce a very high parallelization of the measurements. This method is versatile and can be used to detect and characterize many types of biomolecules from sugars to nucleic acids, proteins, ribosomes and even viral capsids .
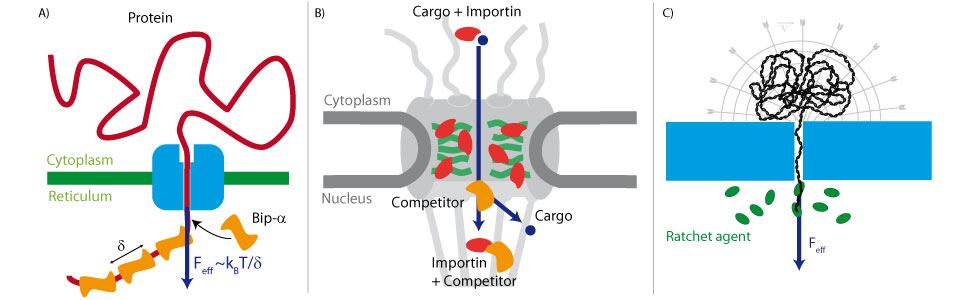
We used this method to measure the energy landscape of translocation for long biomolecules inside nanopores. We have shown that the deGennes-Brochard suction model was an effective description of the hydrodynamical injection of flexible polymers. This model was also extended to the use of electrical field as a driving parameter and also to semi-flexible polymers. The energy landscape of tranlocation can also be modified by grafting artificial polymers that mimicks the unstructured proteins present in the nuclear pore complex. Directionnality can also been induced by using a gradient of ratcheting agents between the two sides of the membrane .
Nuclear pore complex structure and dynamics
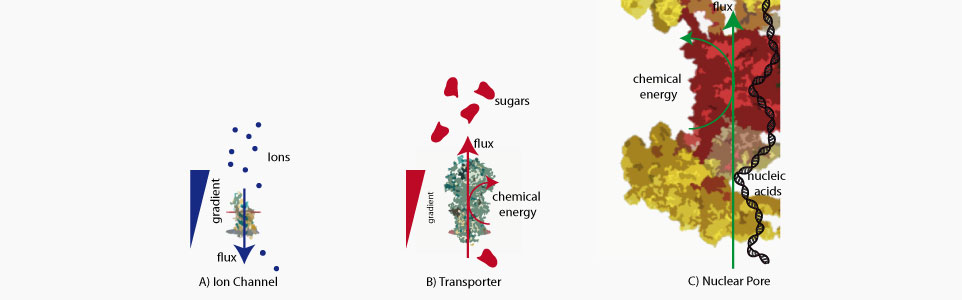
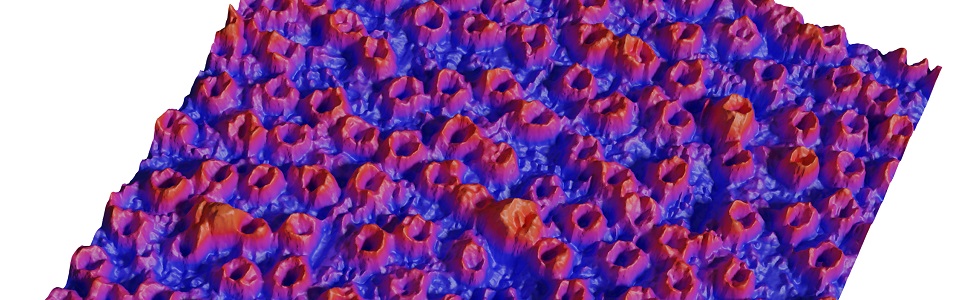
Biological nanopores are uncanny molecular machines that perform a large variety of cellular function ranging from sorting of biomolecules, to building of the cellular osmotic pressure and folding of newly synthetized proteins. Their performance measured through their energetic efficiency, directionnality or selectivity have no equivalent among artificial systems. During the past years we have focused on one of this nanopore called the nuclear pore complex whose consume chemical energy in order to drive the transport of proteins, DNA and RNA inside or outside of the nucleus.
The nuclear pore complex is the only gateway between the cell nucleus and the cytoplasm. It is composed of a large number of components (30 proteins present in multiple copies) including unstructured proteins (FG-nups). The mechanisms of its selectivity and directionnality are still heavily debated. We have used optical super resolution microscopy (dSTORM) to reveal the structural plasticity of this pore during a developmental process. We have also shown and characterized the formation of square lattice organized nanodomains of nuclear pores during this process.