Tools
Create a SEWARD input from a xyz file
This PERL script goes from a XYZ file to a SEWARD file for the MOLCAS software. Atoms are identified by their number in the xyz file. It require the ANO-RCC file to work properly. Basically, it reads each line of the xyz file and stores it. Then for each kind of atoms, it searches the corresponding basis set and the contraction level given. Then it produces a seward file.
WARNING : For some atoms (typically hydrogen and helium), the contraction level found won't be correct as the ANO-RCC file is not written in the same way as for other atoms. The script generates a warning for hydrogen atoms.
Specific keywords such as "symmetry" are to be given manually.
./xyz_to-molcas.pl ECUBEK-sym.xyz DZP
to convert ECUBEK-sym.xyz file to a seward input with a DZP (double zeta polarized) contraction level.
- The script, an input and output file (930 Ko)

Create a CASDI Input from a RASSCF Output
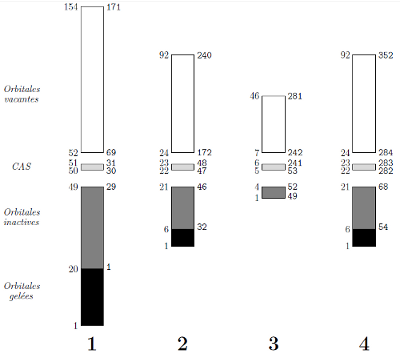
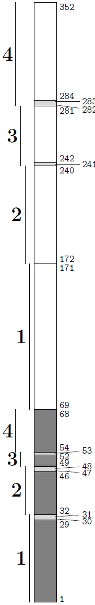
./molcas_ddci.pl phenal-2.in
This PERL script computes the correct numerotation of orbitals for the CASDI package. You have to specify the number of symmetry as well as the number of active electrons and the prefix of the project. In the output file, some fields have to be entered manually.

Compute t and U from a DDCI calculation
./extraction_propre.pl phenal2.coeff
This PERL script computes t and U from the projection of the four first states (three singlet, one triplet) on the model space given by the CAS(2,2) active space. You have to give the projections of the three singlet and the four energies. (singlet first then triplet and finally the two excited singlet).

Create a Tikz picture from a xyz file
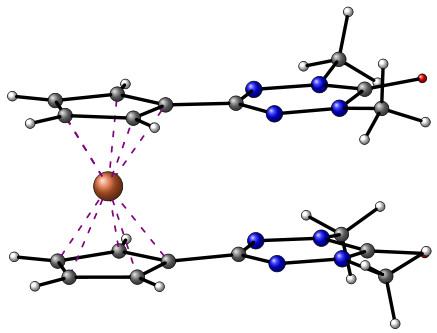
./xyztotikz_advanced.pl PECLIT.xyz 120 84 -38
This PERL script draws a molecule from a xyz file. The view is in cavalier perspective. The first parameter is the filename, the second one is the rotation angle in degrees around the z axis, the third one is the rotation angle around the y axis and the last one around the x axis.
To correctly draw the molecule, atoms are reordered depending on their x coordinate and then the picture is drawn from the background to the foreground with the european convention for xyz axis. You can custom the color or size of atoms playing with the script.
Change coordinates of xyz file
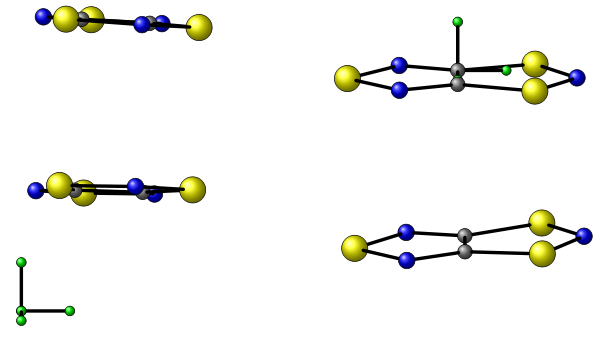
This script performs first a translation so that the first atom of the xyz file is in (0,0,0) the the second atom defines the x axis, the third one define the y plane and consequently the z axis. The picture shows the axes of the input script on the left and the final axes on the right.
ENS Lyon Beamer templates
I created several beamer templates for each color set. Each template contains the different files and a simple compiled example (with pdfLatex). It also includes a fully vectorized pdf logo of the ENS. You can use it to your needs, modify or expand it. All the images were produced with Inkscape and can be edited with this software.
- Blue template(46 ko) Preview
- Brown template(46 ko) Preview
- Green template(46 ko) Preview
- Dark green template(46 ko) Preview
- Multicolor template(46 ko) Preview
- Orange template(46 ko) Preview
- Red template(46 ko) Preview
- Rose template(46 ko) Preview
- Yellow template(46 ko) Preview
- Black & white(46 ko)
- Violet(46 ko)
- All template(390 ko)






