1. scRNASeq technologies
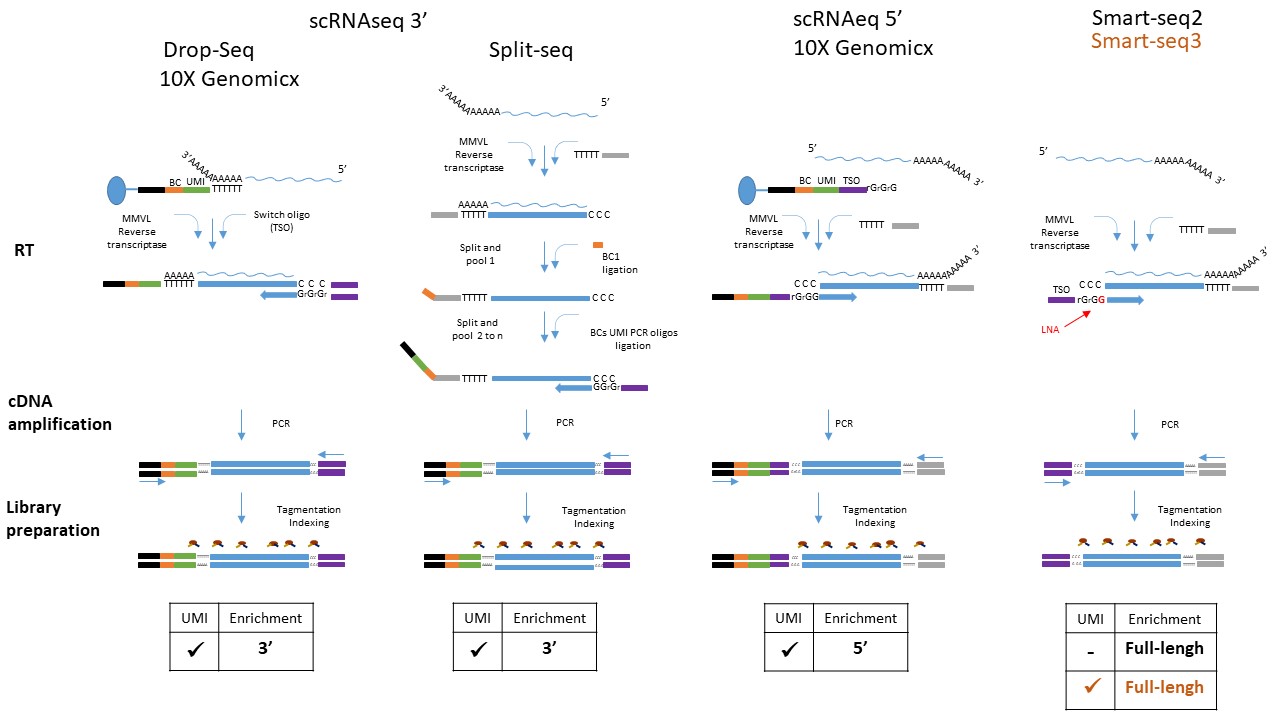
Protocols
The goal of single-cell RNA sequencing is to capture the few RNA molecule with a poly-A tail (\(\sim 10^5\)) in a cell.
Different protocols are available depending on the cell isolation method:
- Cell encapsulation in droplets (10X Genomix Chromium)
- Indexing, or combination on a pool of cells (Split-Seq, Rosenberg et al. 2018)
- Cell sorting (FACS, Fluidigm C1, CellenOne) Smartseq 2/3 Picelli et al. 2014
Given the low amount of material present in each cell, most scRNAseq methods use UMI to prevent PCR amplification bias. UMI or Unique Molecular Identifiers are small barcode that are linked to the 3’ end of mRNA molecules before any amplification step (Islam et al. 2013). This technique allows us to easily remove PCR amplification bias from the data.
Data
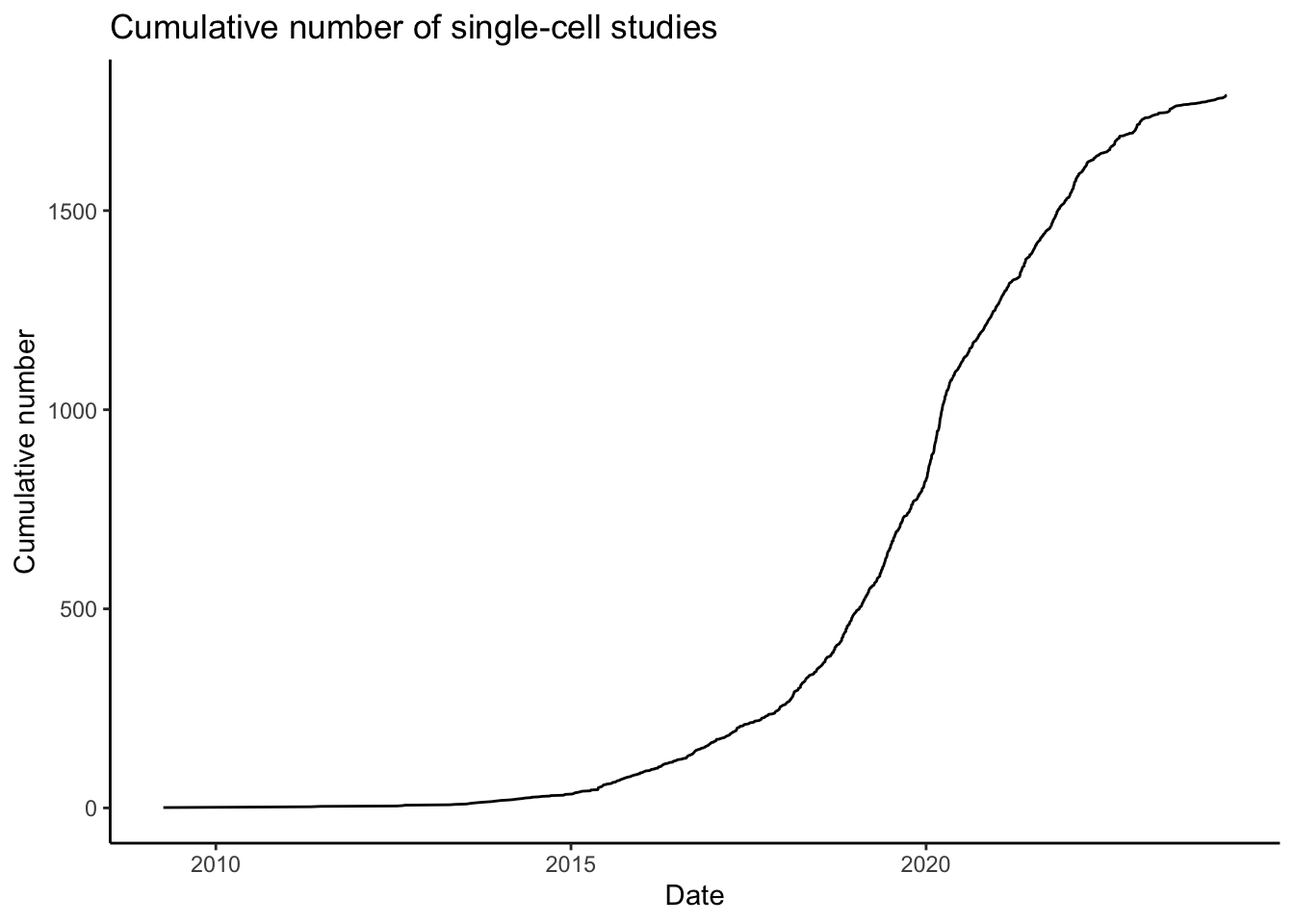
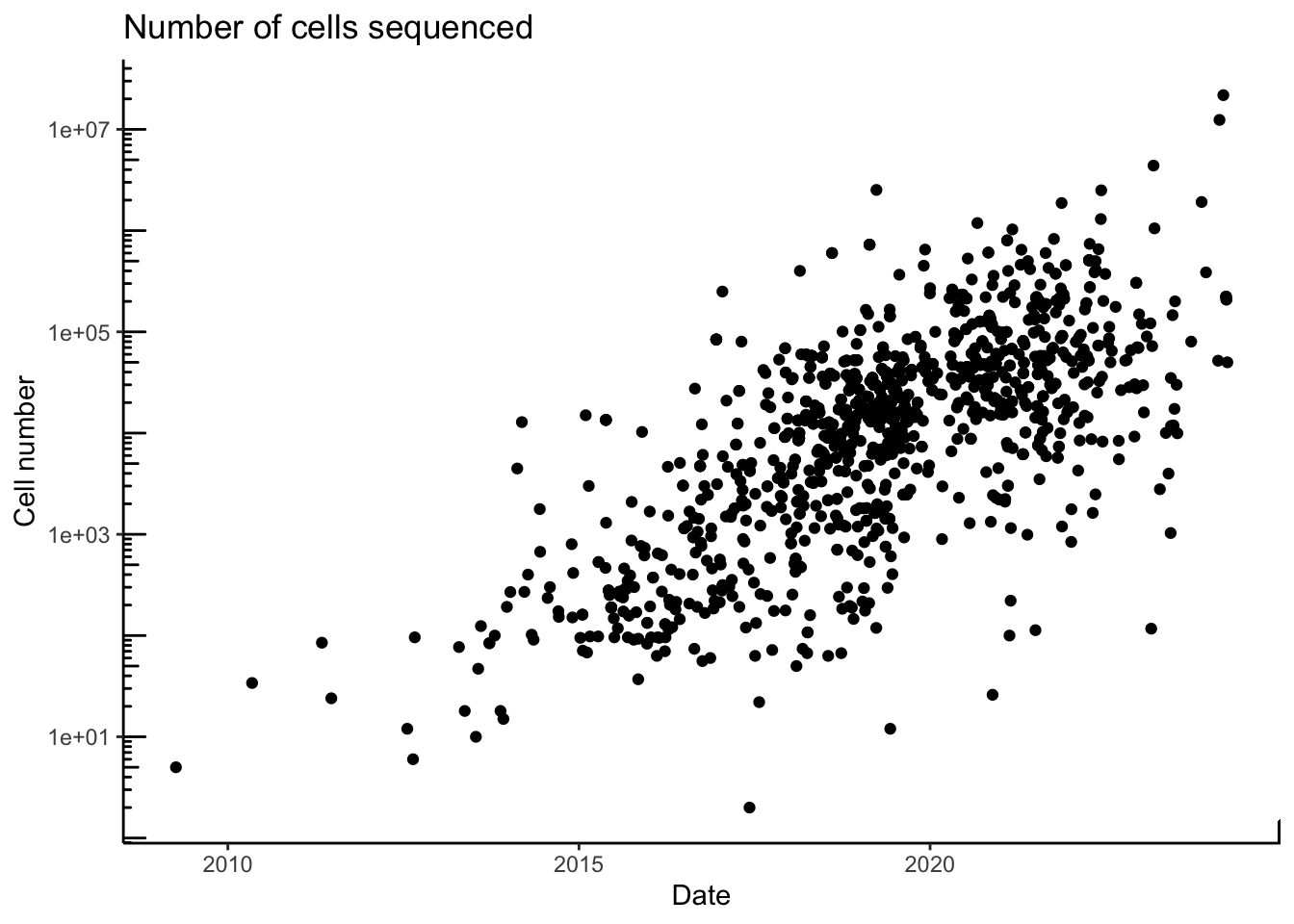
A single-cell experiment can sequence from few hundred to hundreds of thousand individuals cells
The input to a single-cell RNA sequencing (scRNAseq) method is a collection of cells. Formally, the desired output is a transcript or genes (\(M\)) x cells (\(N\)) matrix \(X^{N \times M}\) that describes, for each cell, the abundance of its constituent transcripts or genes. More generally, single-cell genomics methods seek to measure not just transcriptional state, but other modalities in cells, e.g., protein abundances, epigenetic states, cellular morphology, etc.
Ideally, we want a scRNASeq method that:
- is universal in terms of cell size, type and state
- have no minimum input requirements (works on cells with low amount of RNA)
- assay every cell (100% capture rate)
- detect every transcript or gene in every cell (100% sensitivity)
- only detect the cell transcript (100% specificity)
- identify individual transcripts by their full-length sequence
- assign transcripts correctly to cells (no cell doublets)
Analysis
The generation of single-cell RNA-seq data is just the first step in understanding the transcriptomes cells. To interpret the data reads must be aligned or pseudoaligned, Unique Molecular Identifiers (UMIs) counted, and large cell x gene matrices examined. The growth in single-cell RNA-seq analysis tools for these tasks has been breathtaking. The graph below, plotted from real-time data downloaded from the scRNA-seq tools database, shows the number of tools published since 2016.
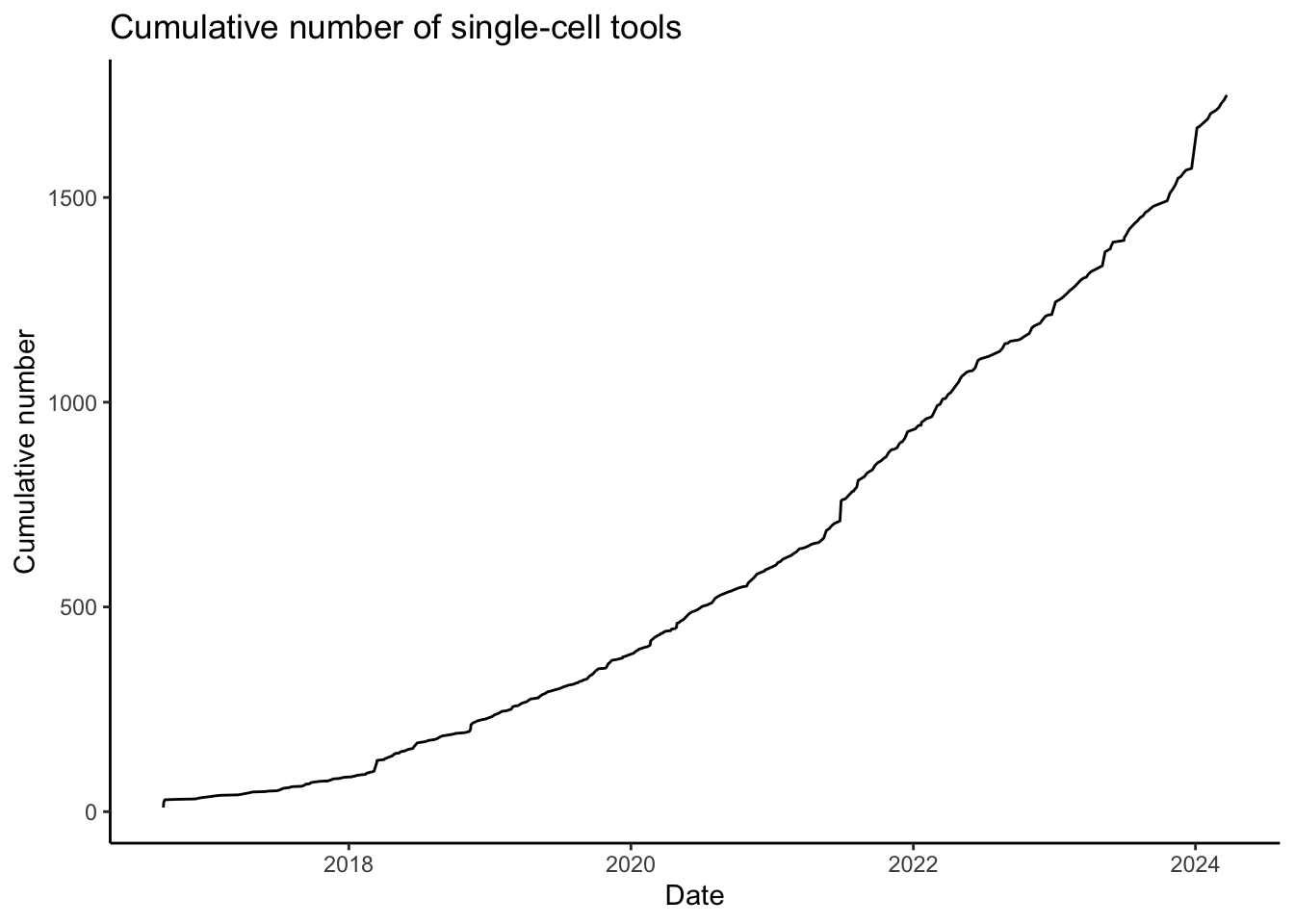
We are not going to learn how to use all these tools, but instead, focus on commonly used ones, to perform every step of a single-cell RNASeq workflow.
In the following all quoted block like this one will correspond to things that you will have to do.
Click on the Next link