Using riverbed with multiple channels
multiple_channels.RmdLoad example datasets
Use two transects s1 and s2 already included as example datasets in the package. They correspond to the same place, at two different times.
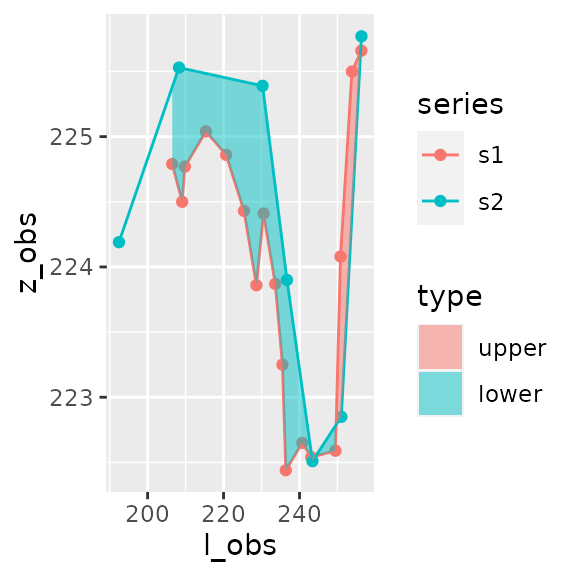
You can compare those two transects and assess the overall change between them through a simple call to area_between().
result_area=area_between(s1,s2)
result_area
#> $data
#> # A tibble: 237 x 13
#> l z1 p z2 a type l_obs z_obs series zmin zmax lmin
#> <dbl> <dbl> <chr> <dbl> <dbl> <fct> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 10.1 228. observ… 228. 0 <NA> 10.1 228. s1 NA NA NA
#> 2 10.1 228. observ… 228. 8.68e-4 upper 10.1 228. s2 NA NA NA
#> 3 10.3 228. inters… 228. NA upper NA NA <NA> NA NA NA
#> 4 10.3 228. inters… 228. -1.13e-1 lower NA NA <NA> NA NA NA
#> 5 12.3 227. observ… 227. 0 <NA> 12.3 227. s1 NA NA NA
#> 6 12.3 227. interp… 227. -2.40e-2 lower NA NA s2 NA NA NA
#> 7 12.7 227. inters… 227. NA lower NA NA <NA> NA NA NA
#> 8 12.7 227. inters… 227. 4.79e-3 upper NA NA <NA> NA NA NA
#> 9 12.9 227. interp… 227. 0 <NA> NA NA s1 NA NA NA
#> 10 12.9 227. observ… 227. 1.14e+0 upper 12.9 227. s2 NA NA NA
#> # … with 227 more rows, and 1 more variable: lmax <dbl>
#>
#> $area
#> [1] -98.07248
#>
#> $area_by_type
#> # A tibble: 2 x 2
#> type area
#> <fct> <dbl>
#> 1 upper 7.76
#> 2 lower -106.
#>
#> $sigma_area
#> [1] 0
plot_area(result_area)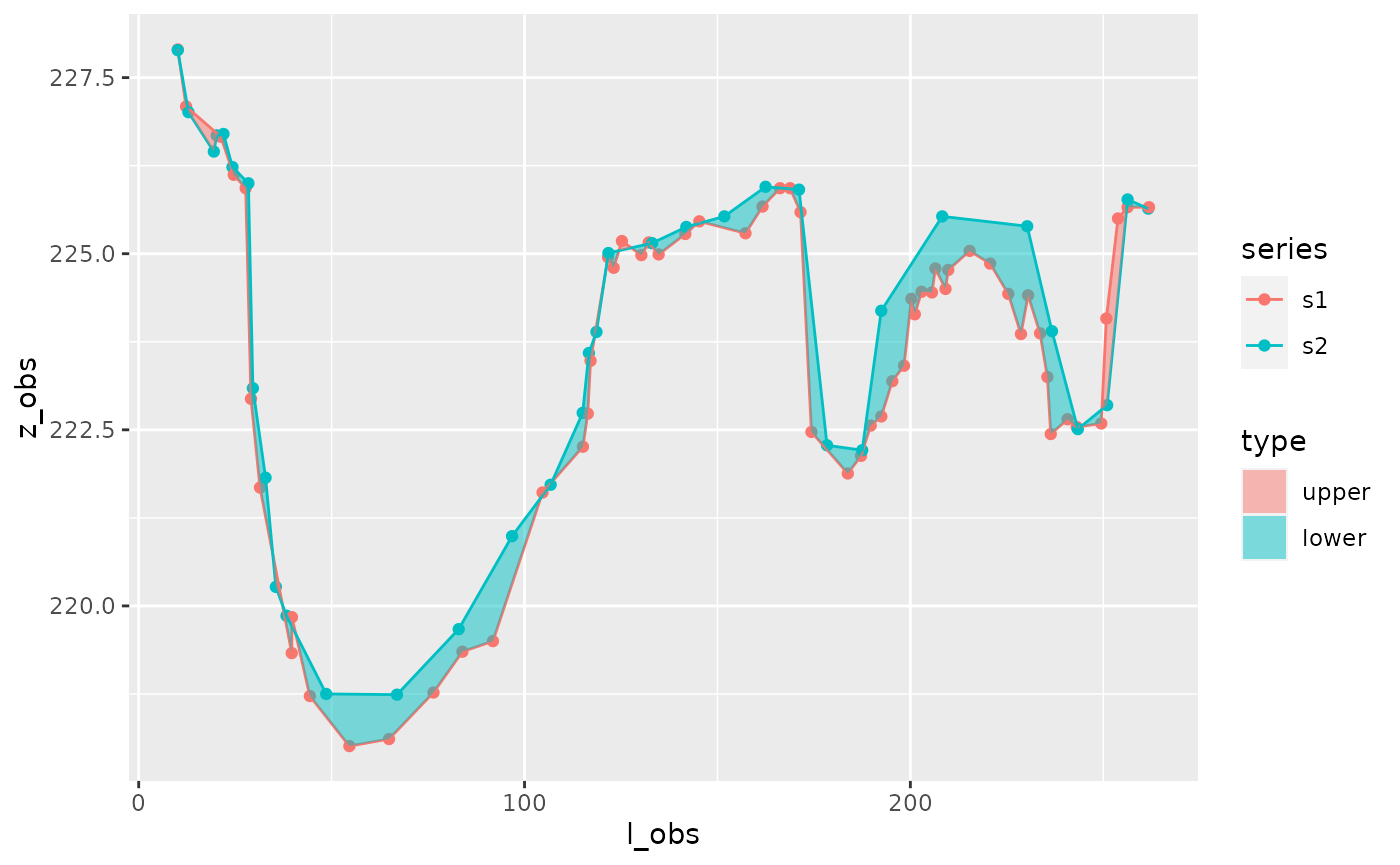
Identify channels with get_channels()
On the other hand, these transects might correspond to a multi-channel riverbed, in which case you might want to assess the changes of each channel individually. It is then necessary to first identify channels.
channels1=get_channels(s1, hmin=1, hmax=5.5)
channels1
#> # A tibble: 3 x 9
#> id_c i_a i_b l_a l_b z_a z_b z_min z_max
#> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 5 20 28.2 122. 225. 225. 218. 225.
#> 2 2 32 40 173. 200. 224. 224. 222. 224.
#> 3 3 50 59 231. 251. 224. 224. 222. 224.
plot_channels(channels1,sr=s1)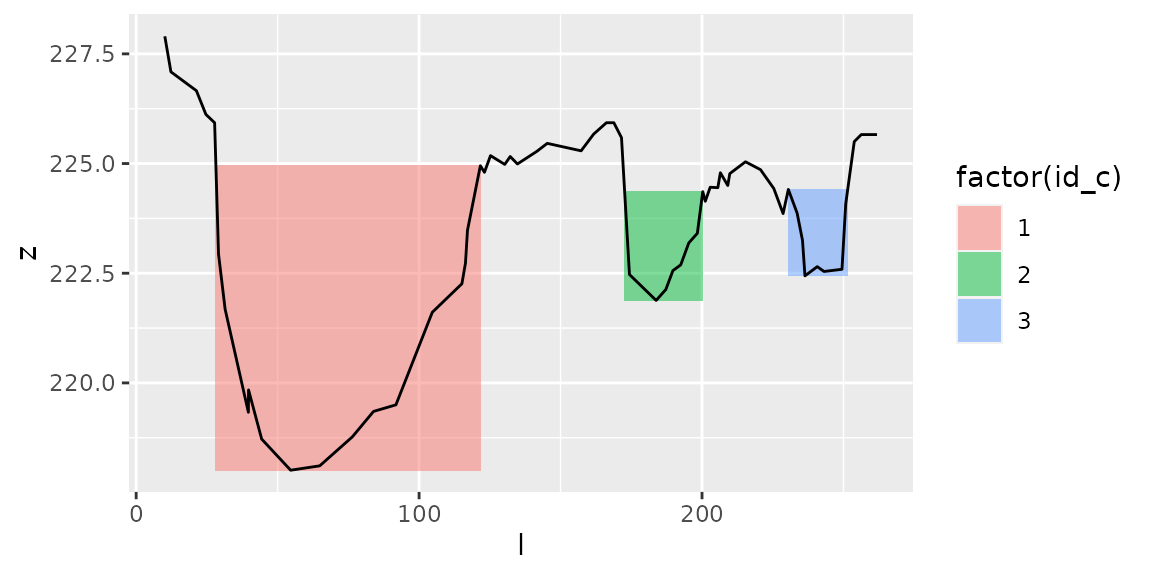
- i_a and i_b: indices of observations delimiting the channel,
- l_a and l_b: longitudinal coordinates of points delimiting the channel
- z_a and z_b: height of points delimiting the channel
- z_min and z_max: minimum and maximum height of the channel-delimited profile
The type of channel returned can be either
- “bankfull” (the default, as above)
- “levee-to-levee” (see example below), which is delimited on both sides by local maxima
Here is what you would get with type=“levee-to-levee”:
channels1=get_channels(s1, hmin=1, hmax=5.5, type="levee-to-levee")
channels1
#> # A tibble: 3 x 9
#> id_c i_a i_b l_a l_b z_a z_b z_min z_max
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 1 20 10.1 122. 228. 225. 218. 228.
#> 2 2 31 40 169. 200. 226. 224. 222. 226.
#> 3 3 51 60 231. 256. 224. 226. 222. 226.
plot_channels(channels1,sr=s1)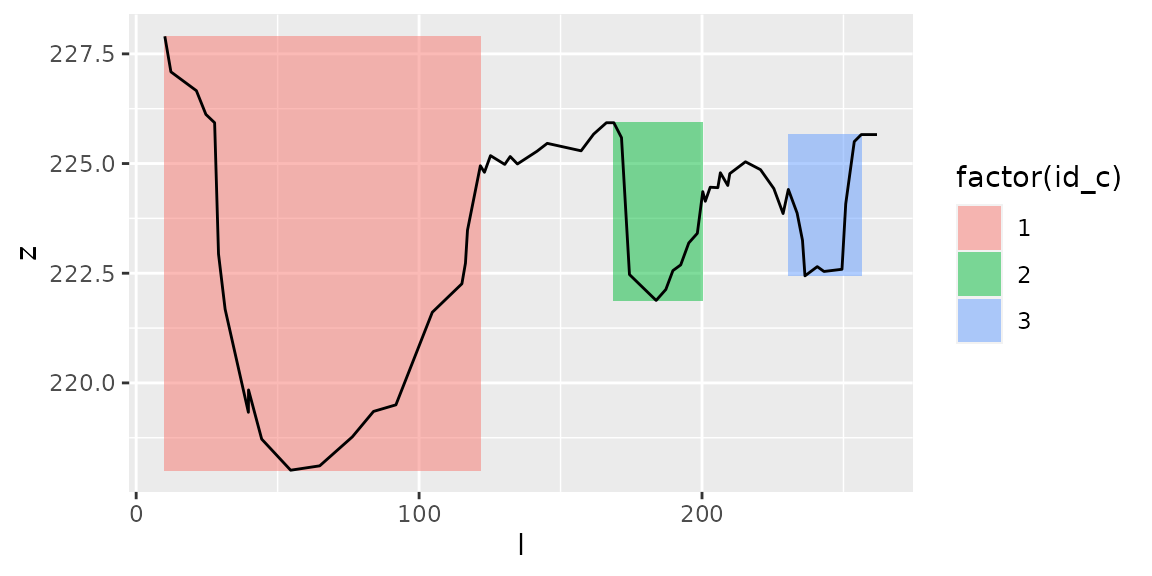
Effect of parameters hmin and hmax
Parameters hmin and hmax
Local minima must be at \(z<z_{min}+h_{max}\) to define a channel’s bottom.
Local maxima must be at \(z>z_{localmin}+h_{min}\) to define a channel’s banks.
Hence \(hmin\) corresponds to the minimum water depth in a channel, and \(hmax\) corresponds to the maximum height of a channel’s bottom.
See the effect of these parameters:
channels1=get_channels(s1, hmin=0.5, hmax=4)
plot_channels(channels1,sr=s1)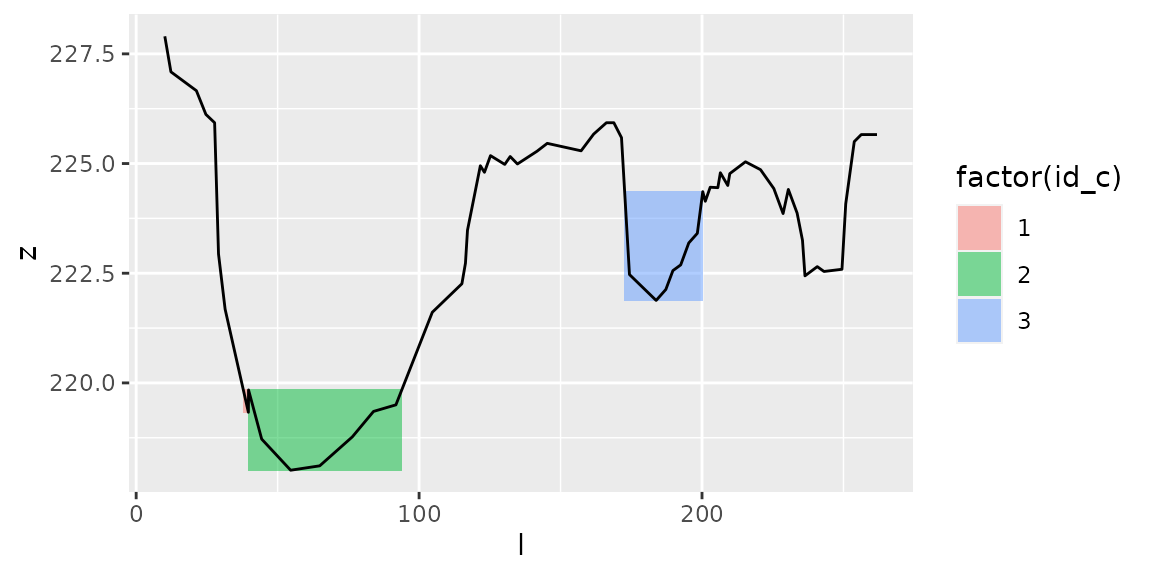
channels1=get_channels(s1, hmin=3, hmax=5)
plot_channels(channels1,sr=s1)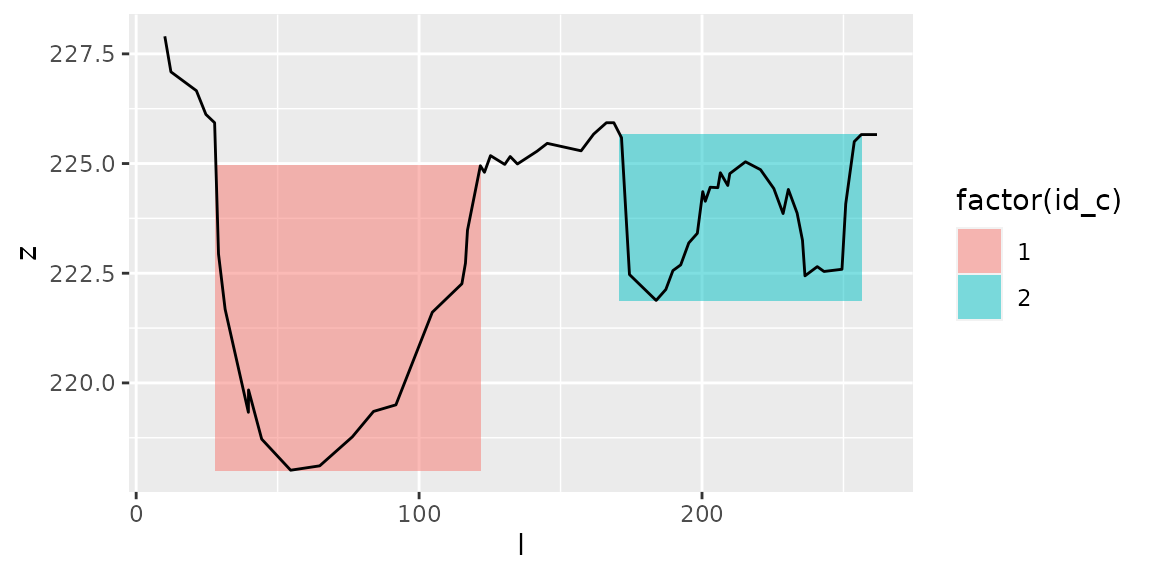 # Regroup channels through dates
# Regroup channels through dates
Here, with the same values \(h_{min}\) and \(h_{max}\), we get 3 distinct channels at both dates:
channels1=get_channels(s1,hmin=1,hmax=5.5)
plot_channels(channels1,sr=s1)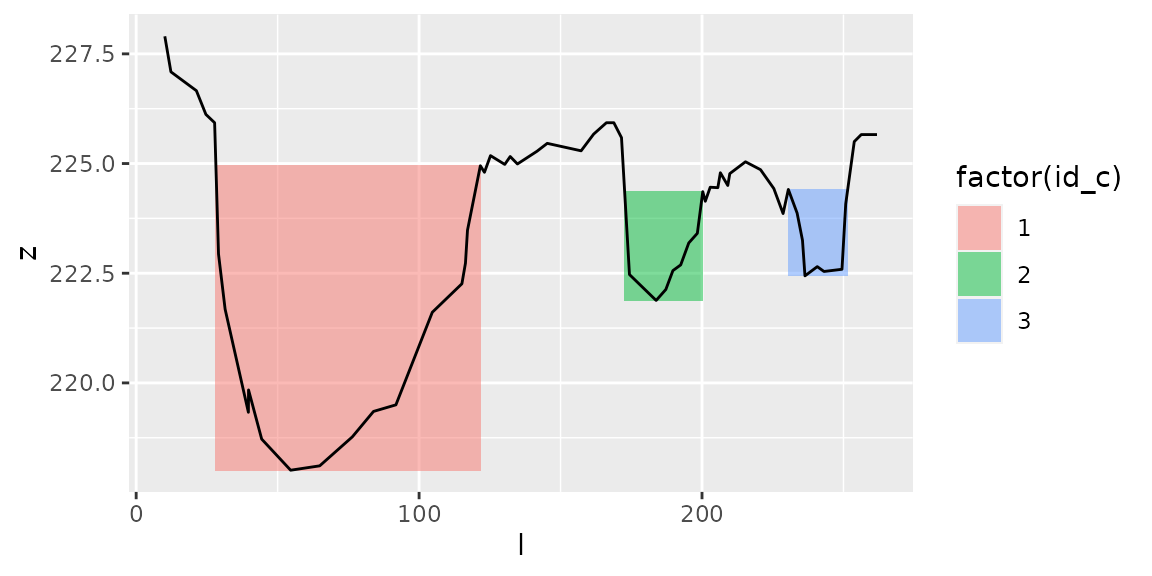
channels2=get_channels(s2,hmin=1,hmax=5.5)
plot_channels(channels2,sr=s2)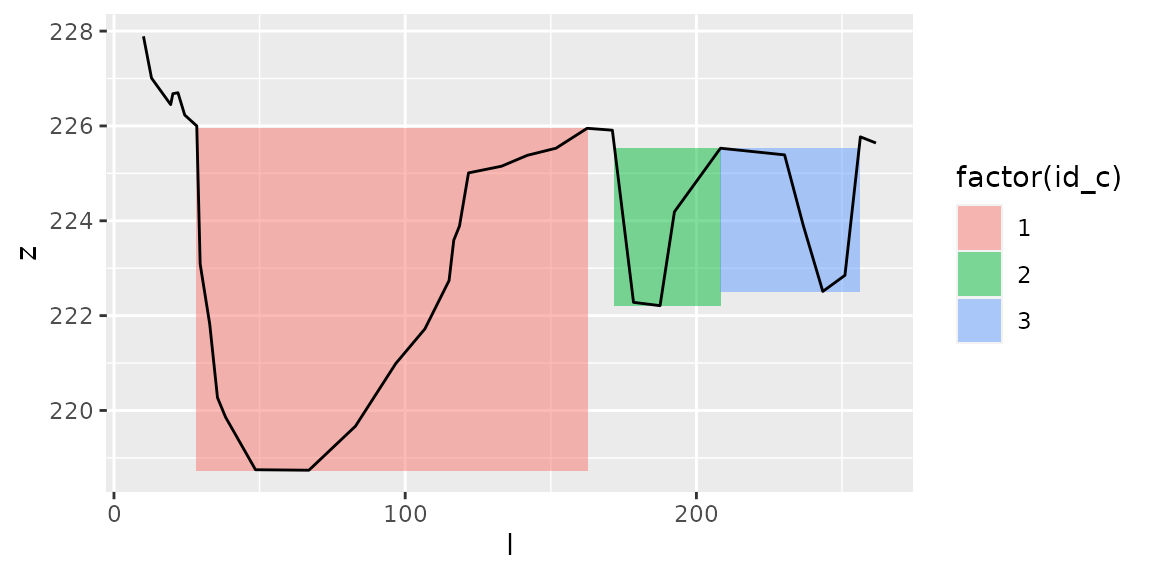
… But these correspond to different \(l\) coordinates.
So, we have to somehow “regroup” these coordinates to be able to compare the transects with channel-wise area calculations.
Example of channel-wise area calculation through two dates
get_channels()
Let’s apply the get_channels() function to all transects (the column identifying those being called “id_transect”).
transects=bind_rows(bind_cols(id_transect="s1",s1),
bind_cols(id_transect="s2",s2))%>%
group_by(id_transect) %>%
tidyr::nest()The table transects gathers the data of both transects s1 and s2. Let’s apply get_channels() iteratively to each transect:
channels =transects %>%
mutate(data=purrr::map(data,get_channels, hmin=1, hmax=4.5)) %>%
tidyr::unnest(cols=data)
channels
#> # A tibble: 6 x 10
#> # Groups: id_transect [2]
#> id_transect id_c i_a i_b l_a l_b z_a z_b z_min z_max
#> <chr> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 s1 1 5 20 28.2 122. 225. 225. 218. 225.
#> 2 s1 2 32 40 173. 200. 224. 224. 222. 224.
#> 3 s1 3 50 59 231. 251. 224. 224. 222. 224.
#> 4 s2 1 7 24 28.5 162. 226. 226. 219. 226.
#> 5 s2 2 25 30 172. 208. 226. 226. 222. 226.
#> 6 s2 3 28 34 208. 256. 226. 226. 223. 226.regroup_channels()
Both transects correspond to 3 channels with identifier id_c (1,2,3).
We regroup these channels with function regroup_channels(), which returns for each channel longitudinal limits l_a and l_b. These limits include the total width of both channels.
regrouped_channels=regroup_channels(channels,by_id=id_c)
regrouped_channels
#> # A tibble: 3 x 3
#> id_c l_a l_b
#> <int> <dbl> <dbl>
#> 1 1 28.2 162.
#> 2 2 172. 208.
#> 3 3 208. 256.
plot_channels(channels1, s1)+
geom_vline(data=regrouped_channels, aes(xintercept=l_a), col="grey")+
geom_vline(data=regrouped_channels, aes(xintercept=l_b), col="grey")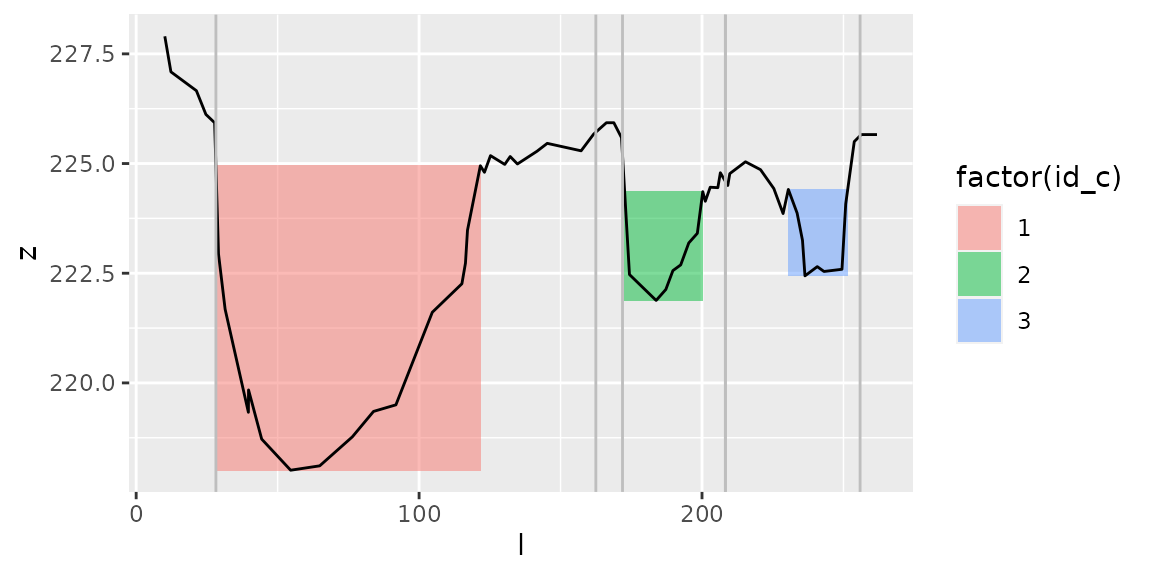
plot_channels(channels2, s2)+
geom_vline(data=regrouped_channels, aes(xintercept=l_a), col="grey")+
geom_vline(data=regrouped_channels, aes(xintercept=l_b), col="grey")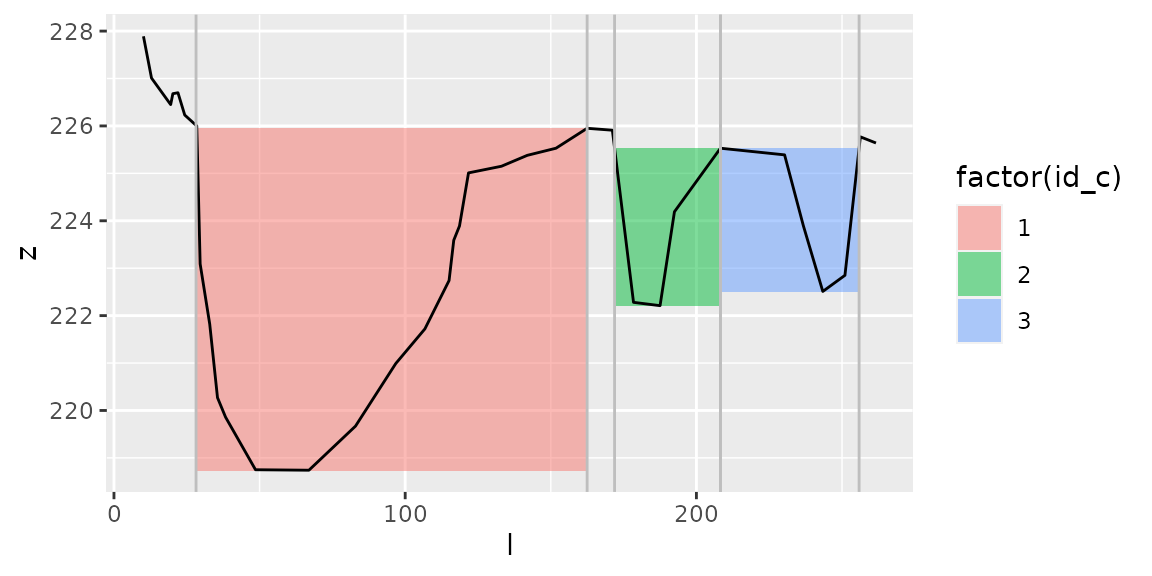 ## area_between()
## area_between()
areas_result=transects %>%
mutate(subdata=purrr::map(data,cut_series,
channels=regrouped_channels)) %>%
tidyr::unnest(cols=subdata) %>%
tidyr::pivot_wider(id_cols=id_c, names_from=id_transect,values_from=sr) %>%
mutate(area_complete=purrr::map2(s1,s2,area_between))
areas_result
#> # A tibble: 3 x 4
#> id_c s1 s2 area_complete
#> <int> <list> <list> <list>
#> 1 1 <tibble [26 × 2]> <tibble [19 × 2]> <named list [4]>
#> 2 2 <tibble [14 × 2]> <tibble [6 × 2]> <named list [4]>
#> 3 3 <tibble [17 × 2]> <tibble [7 × 2]> <named list [4]>format result
areas_formatted_result= areas_result%>%
mutate(area=purrr::map_dbl(area_complete,~.x$area)) %>%
mutate(area_lower=purrr::map_dbl(area_complete,
~.x$area_by_type %>%
filter(type=="lower") %>% pull(area))) %>%
mutate(area_upper=purrr::map_dbl(area_complete,
~.x$area_by_type %>%
filter(type=="upper") %>% pull(area))) %>%
mutate(plot=purrr::map(area_complete,plot_area)) %>%
select(id_c,area,area_lower,area_upper,plot)
areas_formatted_result
#> # A tibble: 3 x 5
#> id_c area area_lower area_upper plot
#> <int> <dbl> <dbl> <dbl> <list>
#> 1 1 -47.8 -48.6 0.800 <gg>
#> 2 2 -27.9 -27.9 0 <gg>
#> 3 3 -23.9 -29.6 5.66 <gg>
areas_formatted_result$plot
#> [[1]]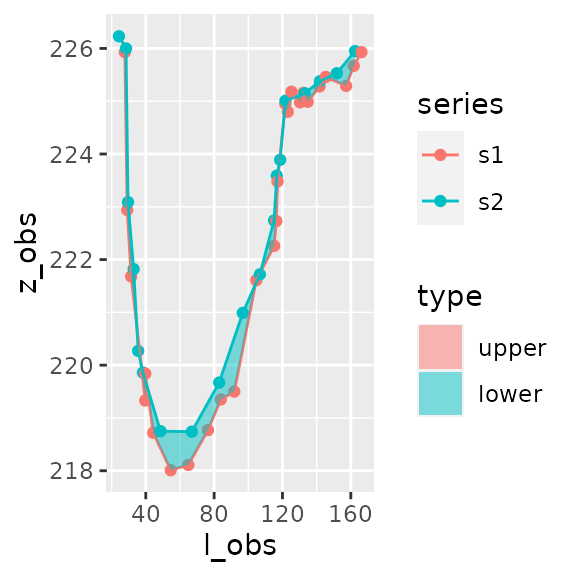
#>
#> [[2]]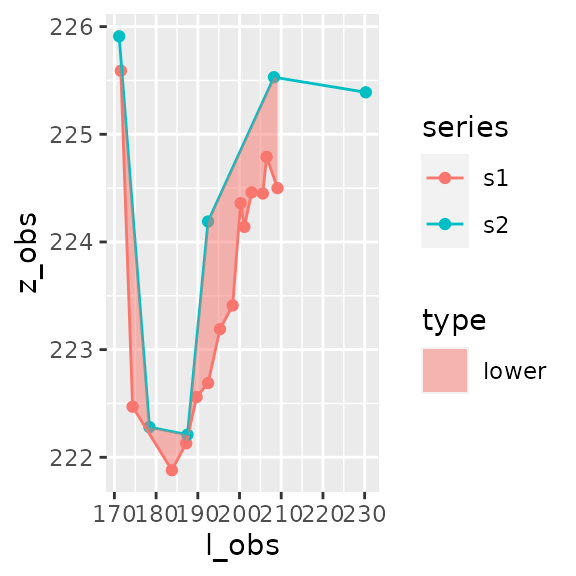
#>
#> [[3]]